Getting started
Install and import sme
[1]:
!pip install -q sme
import sme
from matplotlib import pyplot as plt
import numpy as np
print("sme version:", sme.__version__)
sme version: 1.6.0+d0478c5
Importing a model
to load an existing sme or xml file:
sme.open_file('model_filename.xml')to load a built-in example model:
sme.open_example_model()
[2]:
my_model = sme.open_example_model()
Getting help
to see the type of an object:
type(object)to print a one line description of an object:
repr(object)to print a multi-line description of an object:
print(object)to get help on an object, its methods and properties:
help(object)
[3]:
type(my_model)
[3]:
sme.Model
[4]:
repr(my_model)
[4]:
"<sme.Model named 'Very Simple Model'>"
[5]:
print(my_model)
<sme.Model>
- name: 'Very Simple Model'
- compartments:
- Outside
- Cell
- Nucleus
- membranes:
- Outside <-> Cell
- Cell <-> Nucleus
[6]:
help(my_model)
Help on Model in module sme object:
class Model(pybind11_builtins.pybind11_object)
| the spatial model
|
| Method resolution order:
| Model
| pybind11_builtins.pybind11_object
| builtins.object
|
| Methods defined here:
|
| __init__(...)
| __init__(self: sme.Model, filename: str) -> None
|
| __repr__(...)
| __repr__(self: sme.Model) -> str
|
| __str__(...)
| __str__(self: sme.Model) -> str
|
| export_sbml_file(...)
| export_sbml_file(self: sme.Model, filename: str) -> None
|
|
| exports the model as a spatial SBML file
|
| Args:
| filename (str): the name of the file to create
|
| export_sme_file(...)
| export_sme_file(self: sme.Model, filename: str) -> None
|
|
| exports the model as a sme file
|
| Args:
| filename (str): the name of the file to create
|
| import_geometry_from_image(...)
| import_geometry_from_image(self: sme.Model, filename: str) -> None
|
|
| sets the geometry of each compartment to the corresponding pixels in the supplied geometry image
|
| Note:
| Currently this function assumes that the compartments maintain the same colour
| as they had with the previous geometry image. If the new image does not contain
| pixels of each of these colours, the new model geometry will not be valid.
| The volume of a pixel (in physical units) is also unchanged by this function.
|
| Args:
| filename (str): the name of the geometry image to import
|
| simulate(...)
| simulate(*args, **kwargs)
| Overloaded function.
|
| 1. simulate(self: sme.Model, simulation_time: float, image_interval: float, timeout_seconds: int = 86400, throw_on_timeout: bool = True, simulator_type: sme.SimulatorType = <SimulatorType.Pixel: 1>, continue_existing_simulation: bool = False, return_results: bool = True, n_threads: int = 1) -> std::vector<sme::SimulationResult, std::allocator<sme::SimulationResult> >
|
|
| returns the results of the simulation.
|
| Args:
| simulation_time (float): The length of the simulation in model units of time, e.g. `5.5`
| image_interval (float): The interval between images in model units of time, e.g. `1.1`
| timeout_seconds (int): The maximum time in seconds that the simulation can run for. Default value: 86400 = 1 day.
| throw_on_timeout (bool): Whether to throw an exception on simulation timeout. Default value: `True`.
| simulator_type (sme.SimulatorType): The simulator to use: `sme.SimulatorType.DUNE` or `sme.SimulatorType.Pixel`. Default value: Pixel.
| continue_existing_simulation (bool): Whether to continue the existing simulation, or start a new simulation. Default value: `False`, i.e. any existing simulation results are discarded before doing the simulation.
| return_results (bool): Whether to return the simulation results. Default value: `True`. If `False`, an empty SimulationResultList is returned.
| n_threads(int): Number of cpu threads to use (for Pixel simulations). Default value is 1, 0 means use all available threads.
|
| Returns:
| SimulationResultList: the results of the simulation
|
| Raises:
| RuntimeError: if the simulation times out or fails
|
|
| 2. simulate(self: sme.Model, simulation_times: str, image_intervals: str, timeout_seconds: int = 86400, throw_on_timeout: bool = True, simulator_type: sme.SimulatorType = <SimulatorType.Pixel: 1>, continue_existing_simulation: bool = False, return_results: bool = True, n_threads: int = 1) -> std::vector<sme::SimulationResult, std::allocator<sme::SimulationResult> >
|
|
| returns the results of the simulation.
|
| Args:
| simulation_times (str): The length(s) of the simulation in model units of time as a comma-delimited list, e.g. `"5"`, or `"10;100;20"`
| image_intervals (str): The interval(s) between images in model units of time as a comma-delimited list, e.g. `"1"`, or `"2;10;0.5"`
| timeout_seconds (int): The maximum time in seconds that the simulation can run for. Default value: 86400 = 1 day.
| throw_on_timeout (bool): Whether to throw an exception on simulation timeout. Default value: `true`.
| simulator_type (sme.SimulatorType): The simulator to use: `sme.SimulatorType.DUNE` or `sme.SimulatorType.Pixel`. Default value: Pixel.
| continue_existing_simulation (bool): Whether to continue the existing simulation, or start a new simulation. Default value: `false`, i.e. any existing simulation results are discarded before doing the simulation.
| return_results (bool): Whether to return the simulation results. Default value: `True`. If `False`, an empty SimulationResultList is returned.
| n_threads(int): Number of cpu threads to use (for Pixel simulations). Default value is 1, 0 means use all available threads.
|
| Returns:
| SimulationResultList: the results of the simulation
|
| Raises:
| RuntimeError: if the simulation times out or fails
|
| simulation_results(...)
| simulation_results(self: sme.Model) -> std::vector<sme::SimulationResult, std::allocator<sme::SimulationResult> >
|
|
| returns the simulation results.
|
| Returns:
| SimulationResultList: the simulation results
|
| ----------------------------------------------------------------------
| Readonly properties defined here:
|
| compartment_image
| numpy.ndarray: an image of the compartments in this model
|
| An array of RGB integer values for each voxel in the image of
| the compartments in this model,
| which can be displayed using e.g. ``matplotlib.pyplot.imshow``
|
| Examples:
| the image is a 4d (depth x height x width x 3) array of integers:
|
| >>> import sme
| >>> model = sme.open_example_model()
| >>> type(model.compartment_image)
| <class 'numpy.ndarray'>
| >>> model.compartment_image.dtype
| dtype('uint8')
| >>> model.compartment_image.shape
| (1, 100, 100, 3)
|
| each voxel in the image has a triplet of RGB integer values
| in the range 0-255:
|
| >>> model.compartment_image[0, 34, 36]
| array([144, 97, 193], dtype=uint8)
|
| the first z-slice of the image can be displayed using matplotlib:
|
| >>> import matplotlib.pyplot as plt
| >>> imgplot = plt.imshow(model.compartment_image[0])
|
| compartments
| CompartmentList: the compartments in this model
|
| a list of :class:`Compartment` that can be iterated over,
| or indexed into by name or position in the list.
|
| Examples:
| the list of compartments can be iterated over:
|
| >>> import sme
| >>> model = sme.open_example_model()
| >>> for compartment in model.compartments:
| ... print(compartment.name)
| Outside
| Cell
| Nucleus
|
| or a compartment can be found using its name:
|
| >>> cell = model.compartments["Cell"]
| >>> print(cell.name)
| Cell
|
| or indexed by its position in the list:
|
| >>> last_compartment = model.compartments[-1]
| >>> print(last_compartment.name)
| Nucleus
|
| membranes
| MembraneList: the membranes in this model
|
| a list of :class:`Membrane` that can be iterated over,
| or indexed into by name or position in the list.
|
| Examples:
| the list of membranes can be iterated over:
|
| >>> import sme
| >>> model = sme.open_example_model()
| >>> for membrane in model.membranes:
| ... print(membrane.name)
| Outside <-> Cell
| Cell <-> Nucleus
|
| or a membrane can be found using its name:
|
| >>> outer = model.membranes["Outside <-> Cell"]
| >>> print(outer.name)
| Outside <-> Cell
|
| or indexed by its position in the list:
|
| >>> last_membrane = model.membranes[-1]
| >>> print(last_membrane.name)
| Cell <-> Nucleus
|
| parameters
| ParameterList: the parameters in this model
|
| a list of :class:`Parameter` that can be iterated over,
| or indexed into by name or position in the list.
|
| Examples:
| the list of parameters can be iterated over:
|
| >>> import sme
| >>> model = sme.open_example_model()
| >>> for parameter in model.parameters:
| ... print(parameter.name)
| param
|
| or a parameter can be found using its name:
|
| >>> p = model.parameters["param"]
| >>> print(p.name)
| param
|
| or indexed by its position in the list:
|
| >>> last_param = model.parameters[-1]
| >>> print(last_param.name)
| param
|
| ----------------------------------------------------------------------
| Data descriptors defined here:
|
| name
| str: the name of this model
|
| ----------------------------------------------------------------------
| Static methods inherited from pybind11_builtins.pybind11_object:
|
| __new__(*args, **kwargs) from pybind11_builtins.pybind11_type
| Create and return a new object. See help(type) for accurate signature.
Viewing model contents
the compartments in a model can be accessed as a list:
model.compartmentsthe list can be iterated over, or an item looked up by index or name
other lists of objects, such as species in a compartment, or parameters in a reaction, behave in the same way
Iterating over compartments
[7]:
for compartment in my_model.compartments:
print(repr(compartment))
<sme.Compartment named 'Outside'>
<sme.Compartment named 'Cell'>
<sme.Compartment named 'Nucleus'>
Get compartment by name
[8]:
cell_compartment = my_model.compartments["Cell"]
print(repr(cell_compartment))
<sme.Compartment named 'Cell'>
Get compartment by list index
[9]:
last_compartment = my_model.compartments[-1]
print(repr(last_compartment))
<sme.Compartment named 'Nucleus'>
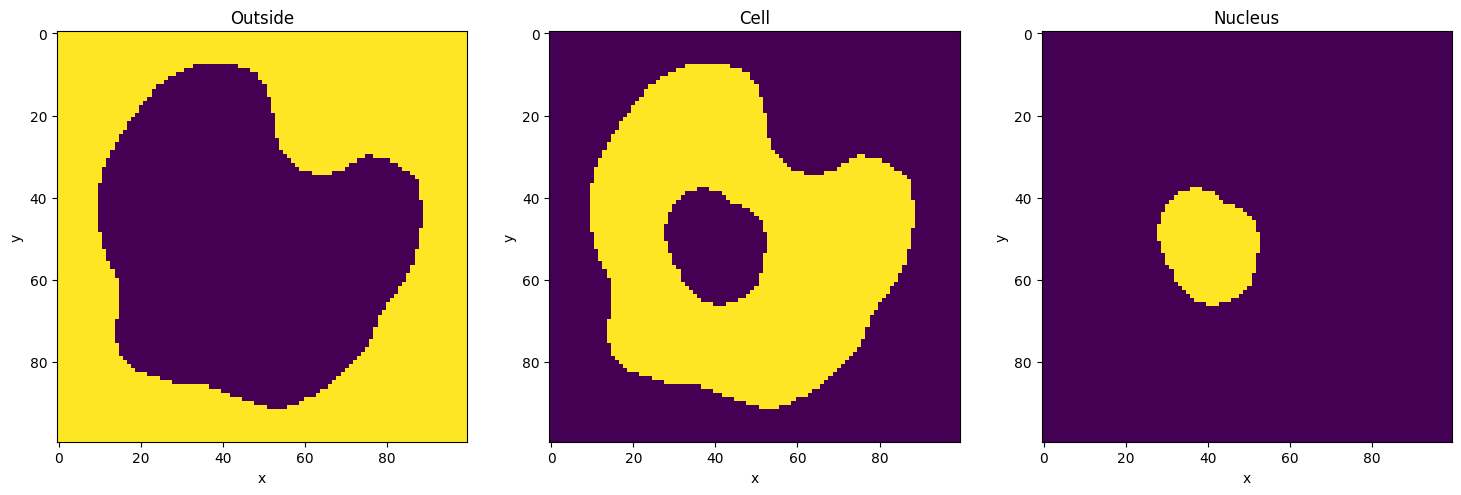
Display geometry of compartments
[10]:
fig, axs = plt.subplots(nrows=1, ncols=len(my_model.compartments), figsize=(18, 12))
for ax, compartment in zip(axs, my_model.compartments):
ax.imshow(compartment.geometry_mask[0], interpolation="none")
ax.set_title(f"{compartment.name}")
ax.set_xlabel("x")
ax.set_ylabel("y")
plt.show()
Display parameter names and values
[11]:
my_reac = my_model.compartments["Nucleus"].reactions["A to B conversion"]
print(my_reac)
for param in my_reac.parameters:
print(param)
<sme.Reaction>
- name: 'A to B conversion'
<sme.ReactionParameter>
- name: 'k1'
- value: '0.3'
Editing model contents
Parameter values and object names can be changed by assigning new values to them
Names
[12]:
print(f"Model name: {my_model.name}")
Model name: Very Simple Model
[13]:
my_model.name = "New model name!"
print(f"Model name: {my_model.name}")
Model name: New model name!
Model Parameters
[14]:
param = my_model.parameters[0]
print(f"{param.name} = {param.value}")
param = 1
[15]:
param.value = "2.5"
print(f"{param.name} = {param.value}")
param = 2.5
Reaction Parameters
[16]:
k1 = my_model.compartments["Nucleus"].reactions["A to B conversion"].parameters["k1"]
print(f"{k1.name} = {k1.value}")
k1 = 0.3
[17]:
k1.value = 0.72
print(f"{k1.name} = {k1.value}")
k1 = 0.72
Species Initial Concentrations
can be Uniform (
float), Analytic (str) or Image (np.ndarray)
[18]:
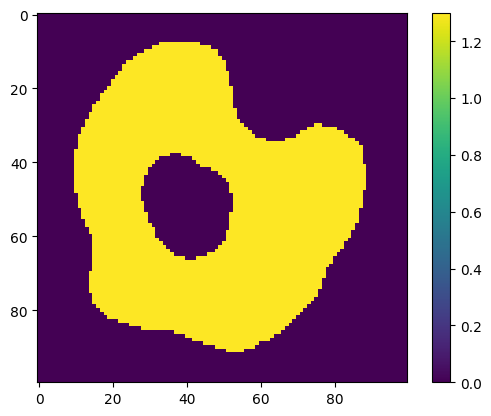
species = my_model.compartments["Cell"].species[0]
print(
f"Species '{species.name}' has initial concentration of type '{species.concentration_type}', value '{species.uniform_concentration}'"
)
species.uniform_concentration = 1.3
print(
f"Species '{species.name}' has initial concentration of type '{species.concentration_type}', value '{species.uniform_concentration}'"
)
plt.imshow(species.concentration_image[0])
plt.colorbar()
plt.show()
Species 'A_cell' has initial concentration of type 'ConcentrationType.Uniform', value '0.0'
Species 'A_cell' has initial concentration of type 'ConcentrationType.Uniform', value '1.3'
[19]:
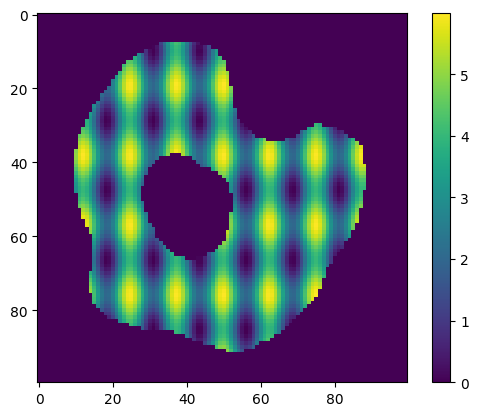
species.analytic_concentration = "3 + 2*cos(x/2)+sin(y/3)"
print(
f"Species '{species.name}' has initial concentration of type '{species.concentration_type}', expression '{species.analytic_concentration}'"
)
plt.imshow(species.concentration_image[0])
plt.colorbar()
plt.show()
Species 'A_cell' has initial concentration of type 'ConcentrationType.Analytic', expression '3 + 2 * cos(x / 2) + sin(y / 3)'
[20]:
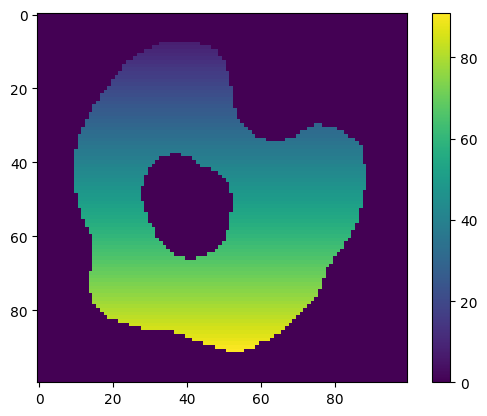
# generate concentration image with concentration = x + y
new_concentration_image = np.zeros(species.concentration_image.shape)
for index, _ in np.ndenumerate(new_concentration_image):
new_concentration_image[index] = index[0] + index[1]
species.concentration_image = new_concentration_image
print(
f"Species '{species.name}' has initial concentration of type '{species.concentration_type}'"
)
plt.imshow(species.concentration_image[0])
plt.colorbar()
plt.show()
Species 'A_cell' has initial concentration of type 'ConcentrationType.Image'
Exporting a model
to save the model, including any simulation results:
model.export_sme_file('model_filename.sme')to export the model as an SBML file (no simulation results):
model.export_sbml_file('model_filename.xml')
[21]:
my_model.export_sme_file("model.sme")
[ ]: