Pixel simulator
Pixel is an alternative PDE solver which uses the simple FTCS method to solve the PDE using the pixels of the geometry image as the grid.
Simulation options
The default settings should work well in most cases, but if desired they can be adjusted by going to Advanced->Simulation options
The simulation options that can be used to fine tune-the Pixel solver.
- Integrator
the explicit Runge-Kutta integration scheme used for time integration
default: 2nd order Heun scheme, with embedded 1st order error estimate
a higher order scheme may be more efficient if the maximum allowed error is very small
see the Time integration section for more information on the integrators
- Max relative local error
the maximum relative error allowed on the concentration of any species at any pixel
default: 0.005
local means the estimated error for a single timestep, at a single point
relative means each error estimate is divided by the species concentration
making this number smaller makes the simulation more accurate, but slower
- Max absolute local error
the maximum error allowed on the concentration of any species at any pixel
default: infinite
local means the estimated error for a single timestep, at a single point
absolute means the error estimate is not normalised by the species concentration
making this number smaller makes the simulation more accurate, but slower
- Max timestep
the maximum allowed timestep
default: infinite
- Multithreading
if enabled, multiple CPU threads can be used
default: disabled
enabling this can make simulations of large models run faster
however it can also make small models run slower
- Max CPU threads
limit the maximum number of CPU threads to be used
default: unlimited
- Symbolic optimization
factor out common subexpressions when constructing the reaction terms
default: enabled
- Compiler optimization level
how much optimization is done when compiling the reaction terms
default: 3
Spatial discretization
Space is discretized using a linear grid with spacing \(\delta x\), \(\delta y\), \(\delta z\). The concentration is defined as a 3d array of values \(c_{i,j,k}\), where the value with index \((i,j,k)\) corresponds to the concentration at the spatial point \((x = i \delta x, y = j \delta y, z = k \delta z)\).
The Laplacian is approximated on this grid using a central difference scheme
which has \(\mathcal{O}(\delta x^2)+\mathcal{O}(\delta y^2)+\mathcal{O}(\delta z^2)\) discretisation errors. Inserting this approximation into the reaction-diffusion equation converts the PDE into a system of coupled ODEs.
Time integration
Time integration is performed using explicit Runge-Kutta integrators. Compared to implicit integrators, they are easier to implement and offer better performance (for the same timestep). However they become unstable if the timestep \(h\) is made too large, so in practice they can end up being slower than implicit methods for stiff problems, where the timestep is forced to be very small to maintain stability.
Integrators differ in their:
order of truncation error
order of embedded error estimate (if any)
number of stages (i.e. cost of a step)
region of stability (can be increased by adding more stages)
memory requirements
Implemented integrators:
- Euler
1st order solution
no error estimate
1 stage
- Embedded Heun / modified Euler
2nd order solution
1st order error estimate
2 stages
see e.g. eq (2.15) of https://doi.org/10.1016/0021-9991(88)90177-5
- Embedded Shu-Osher
3rd order solution
2nd order error estimate
3 stages
see eq (2.17) of https://doi.org/10.1016/0021-9991(88)90177-5
- RK4(3)5[3S*]
4th order solution
3rd order error estimate
5 stages
see alg.6 & tab.6 of https://doi.org/10.1016/j.jcp.2009.11.006
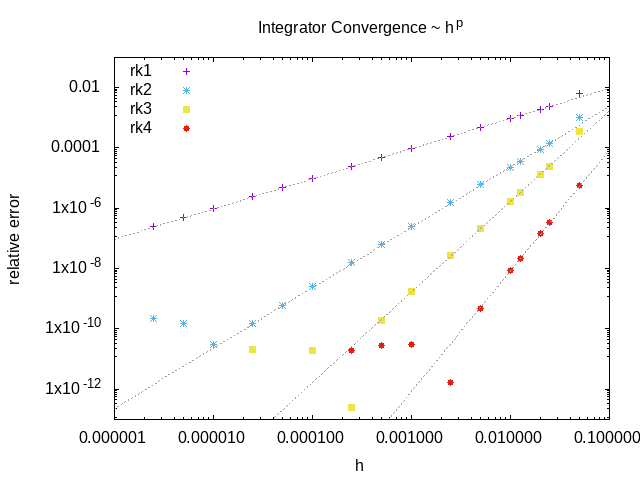
An example of the convergence of the included RK integrators: relative error of the solution at a particular pixel as a function of the stepsize.
These integrators have three sources of error:
- Round-off error due to finite precision
mostly only relevant for high order solvers: not relevant here
- Truncation error due to finite order of integration scheme
we are generally forced by the diffusion term to make the timestep small to maintain stability
also no benefit from making the time integration errors significantly smaller than the spatial discretisation errors
so this is also typically not a concern
- Numerical instability of integrator
a problem when ODEs become stiff, e.g. high rate of diffusion, stiff reaction terms
avoiding these instabilities is our main concern
Adaptive timestep
We use the embedded lower order solution to estimate the error at each timestep, and use this to adapt the stepsize during the integration:
RK gives us a pair of \(u_{n+1}^{(p)} = u_{n} + \mathcal{O}(h^{p+1})\) solutions
difference between \(p, p-1\) solutions gives local error of order \(\mathcal{O}(p)\)
to get the relative error we divide this by \(c = ( |c_{n+1}| + |c_{n}| + \epsilon)/2\)
we use the average of the old and new concentration, plus a small constant, to avoid dividing by zero
we do this for all species, compartments and spatial points, and take the maximum value
if this error is larger than the desired value, the whole step is discarded
the new timestep is given by \(0.98 dt_{old} (err_{desired}/err_{measured})^{1/p}\)
the 0.98 factor is slightly less than 1 to account for the higher order terms that are neglected here
it is better to have a slightly smaller timestep than to have to repeat the whole step
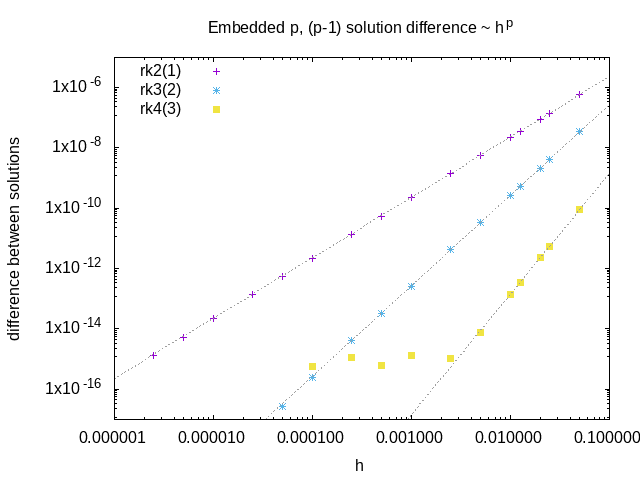
An example of the difference between order p and order p-1 solutions from embedded schemes as a function of the stepsize. This quantity is a measure of the local integration error, and scales like \(h^p\)
Maximum timestep
For the Euler method, we don’t have an embedded lower order solution from which we can estimate of the error, so we can’t automatically adjust the stepsize. However, if we ignore the reaction terms, there is an analytic upper bound on the size of timestep that can be used for Euler (see p125 of https://doi.org/10.1017/CBO9780511781438 ), above which the system becomes unstable:
So if the user selects a timestep larger than this, the simulator automatically reduces it to the above value to avoid the system becoming unstable. Note that the system can still become unstable if the reaction terms are stiffer than the diffusion terms.
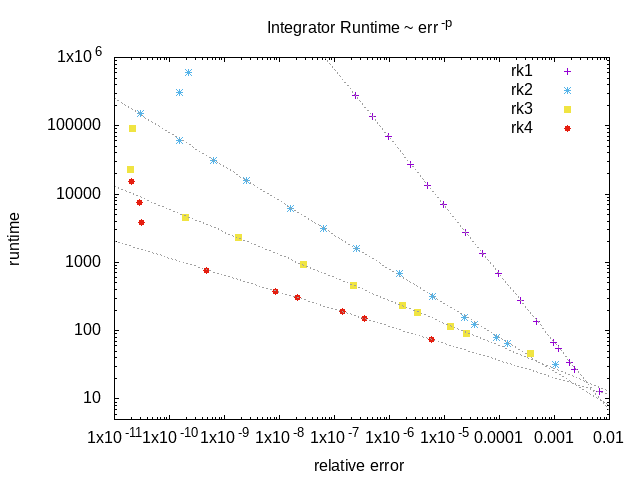
An example of the runtime of the RK integrators as a function of the relative error on the final solution. The higher order integrators offer better performance if a very accurate solution is required, but at lower accuracy the lower order integrators are much faster.
Boundary Conditions
The boundary condition for all boundaries is the “zero-flux” Neumann boundary condition. This is implemented in the spatial discretization by setting the concentration in the neighbouring pixel that lies outside the compartment boundary to be equal to the concentration in the boundary pixel value, or equivalently by setting the neighbour of each boundary pixel to itself.
Compartments
Each compartment is discretized, with the above boundary conditions applied for the diffusion term.
Membranes
Reactions that take place between two compartments involve a flux across the membrane separating the two compartments. For each neighbouring pair of pixels from the two compartments, whose common boundary constitutes the membrane, the flux term is converted into a reaction term that creates or destroys the appropriate amount of species concentration in each pixel.
Non-spatial species
A species can be ‘non-spatial’, which means that at each timestep, its time derivative is calculated as normal at each point in the compartment, but is then spatially averaged over the whole compartment. This can be used to approximate a species with a very high diffusion constant without requiring a correspondingly tiny timestep to maintain the stability of the solver.